United States (EN)
Select your region or country.
Single-molecule fluorescence observation of cells using ORCA®-Quest
The Cell Biophysics Laboratory of the Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System, Gifu University, aims to elucidate the molecular mechanisms inside cells and on cell membranes. In 2022, this laboratory introduced the ORCA-Quest camera for single-molecule fluorescence observation.
We interviewed Professor Kenichi Suzuki and Researcher Koichiro Hirosawa of this laboratory, as well as Rinshi Kasai, a former member of the laboratory until May 2023 and currently the Unit Head of the Division of Advanced Bioimaging at the National Cancer Center Research Institute, regarding the motivations for introducing the ORCA-Quest, their experiences using it, and the perspectives for future research directions.
Current research
The Cell Biophysics Laboratory at the Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System, Gifu University, aims to elucidate the molecular mechanisms by observing individual molecules of proteins, lipids, and other cellular components, obtaining statistic data on event time and frequency.
To elucidate molecular mechanisms, it is essential to observe their behaviors by microscopy. We use ultra-sensitive cameras to capture molecular images, adjusting between high-speed and high-resolution settings as dictated by the experimental requirements, to gain insights into molecular dynamics and interactions. In addition, we are also developing optimal optical systems and imaging techniques to enhance such observations.

Dr. Rinshi Kasai (left), Prof. Kenichi Suzuki (center), Dr. Koichiro Hirosawa (right)
Problems in single-molecule fluorescence observation
The biggest problem in single-molecule fluorescence observation is that the signal emitted from the molecule is very weak. In addition, several factors complicate this process, such as the necessity for high-speed imaging to capture molecular behaviors, which reduces the incident signal per frame. Weak signals can also become obscure by background noise in conventional epi-illumination observations, further complicating signal detection. To reduce background interference, total internal reflection fluorescence (TIRF) microscopy is commonly used in single-molecule fluorescence studies. However, it is also important to reduce the readout noise of the camera, which ultimately outputs the signal as data.
The common approach to address this issue is to use an EM-CCD camera, which detects weak signals by increasing their magnification, or to combine an I.I. (image intensifier) with an sCMOS (scientific CMOS) camera for imaging. However, both EM-CCD cameras and I.I. cameras have the disadvantage that the signal fluctuation is generated during signal multiplication, resulting in low signal quantitativity and resolution. Our laboratory has recently been performing single-molecule super-resolution imaging. Because the reduced resolution due to fluctuations leads to a reduction in the quality of super-resolution images, we required a camera that can detect signals without reliance on I.I. or other methods.


Deciding factor in introducing ORCA-Quest

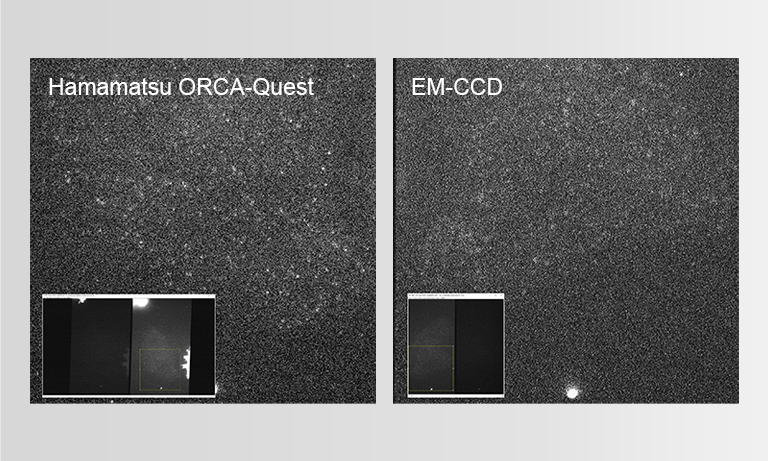
The reason why we decided to implement the ORCA-Quest was the significant improvement in image quality due to reduced noise, which became immediately evident upon reviewing the demonstration images.
Although we knew from the spec sheet that the noise levels were reduced, observing the actual output images and instantly perceiving the remarkable improvement in performance underscored the significant advancements in camera technology.
In our laboratory, we also use the ORCA-Flash4.0, ORCA-Fusion, and other cameras before the ORCA-Quest. While these cameras possess high sensitivity, at the frame rates we require, the signal from a single molecule is too weak to yield satisfactory images with the camera alone. Therefore, we had to combine these sCMOS cameras with I.I. to capture adequate images. However, with ORCA-Quest, we achieved satisfactory single-molecule fluorescence images solely with the camera. This led us to adopt the ORCA-Quest, as we anticipate it would enhance the quality of single-molecule tracking and super-resolution imaging.
Currently, we use both the ORCA-Quest and the I.I.+ previous sCMOS camera setup, selected based on the experimental requirements. For imaging that demands very high speed, the I.I.+sCMOS camera combination may offer greater sensitivity, but in most cases, we have been able to obtain satisfactory images with the ORCA-Quest.
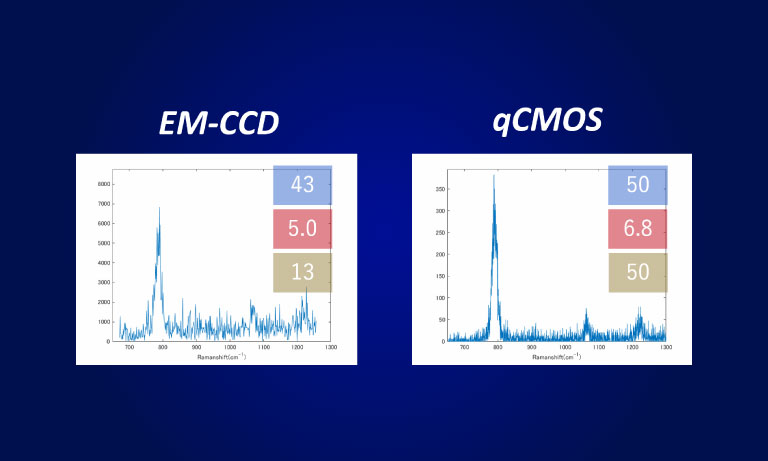
Imaging example
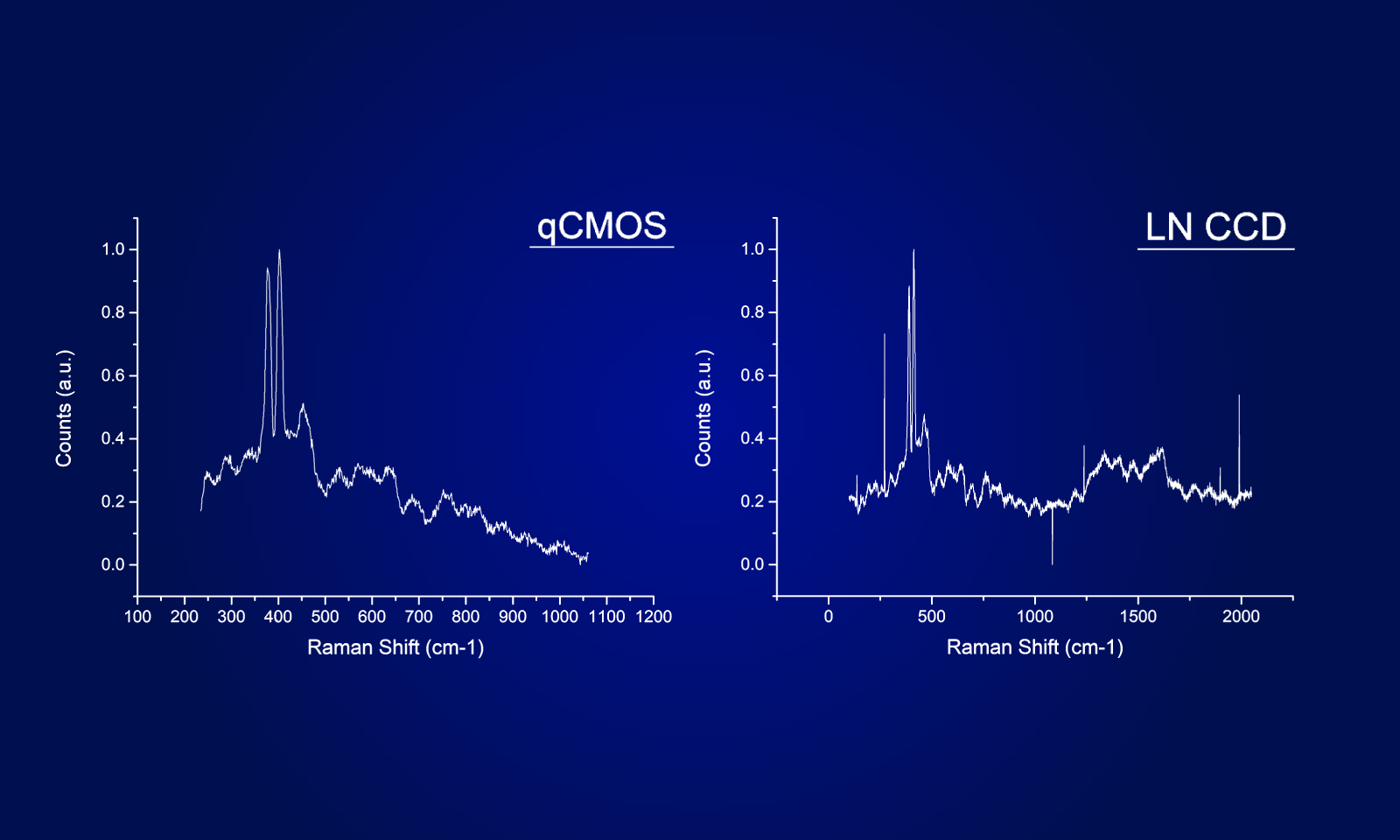
The chemokine receptor CXCR4, labeled with the fluorescent protein mStayGold, was expressed in CHO cells derived from Chinese hamster ovaries, and single-molecule fluorescence observation was performed using the ORCA-Quest.
Scan mode:Standard scan
Frame rate:40 frames/sec(1024 × 1024)
Scan mode:Ultra-quiet scan
Frame rate:40 frames/sec(512 × 512)
Data courtesy of : Dr. Rinshi Kasai, Division of Advanced Bioimaging, National Cancer Center Research Institute
Prospects for future research
As for prospects for future research, we would like to develop our research mainly by using the following two imaging techniques.
1. Multi-wavelength imaging
2. Three-dimensional super-resolution observation
Regarding multi-wavelength imaging, we currently use two wavelengths to observe single-molecule fluorescence. However, we aspire to expand this capability to three or even four wavelengths. For example, if simultaneous three-wavelength imaging becomes feasible, we could observe molecular behaviors at two wavelengths while capturing super-resolution images at the third. This advancement would enable the analysis of molecular interactions and behaviors in even more complex systems than is presently achievable.
Regarding three-dimensional super-resolution observation, our focus has primarily been on monitoring molecular behavior on the cell membrane surface. However, by simultaneously imaging the interior of the membrane with three-dimensional super-resolution observation, we can observe molecular interactions with the endoplasmic reticulum and Golgi apparatus located within. This approach has the potential to elucidate how the endoplasmic reticulum and Golgi apparatus are involved in phenomena occurring on the membrane surface and to clarify how information from events occurring on the membrane surface is transmitted to the inside of the membrane.
There are several methods to capture three-dimensional super-resolution images, but our laboratory adopts a method that uses a cylindrical lens. To acquire a three-dimensional super-resolution image using this method, it is necessary to back-calculate the Z-axis positional information of molecules from their shapes in the two-dimensional image. While the use of an I.I. improves sensitivity, the resolution is reduced, blurring molecular shapes and degrading image quality in super-resolution imaging. The ORCA-Quest addresses this issue by eliminating the need for I.I., thereby preserving image clarity and resolution.
We believe that the ORCA-Quest is a camera poised to significantly advance our research development.

Division of Advanced Bioimaging, National Cancer Center Research Institute
In June 2023, the National Cancer Center Research Institute established Division of Advanced Bioimaging with Prof. Kenichi Suzuki as Chief and Dr. Rinshi Kasai as Laboratory Head.
In this laboratory, the microscopy system identical to that of the Cell Biophysics Laboratory at the Institute for Glyco-core Research (iGCORE) will be used to elucidate signal regulation and signal transduction mechanisms in cancer cells, mainly by single-molecule fluorescence observation.
Researcher profiles

Prof. Kenichi G. N. Suzuki
Cell Biophysics Laboratory of the Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System, Gifu University: Professor.
Division of Advanced Bioimaging, National Cancer Center Research Institute: Chief.
Jan. 1997
Department of Synthetic and Biological Chemistry, Graduate School of Engineering, Kyoto University, Ph. D.
Nov. 1996
Research Associate in Michael P. Sheetz Laboratory, Duke University Medical Center, Department of Cell Biology
Feb. 1999
Researcher in ERATO Kusumi Membrane Organizer Project
Apr. 2005
Research Assistant Professor, Kyoto University, Institute for Frontier Medical Sciences
Oct. 2008
PRESTO Researcher, Japan Science and Technology Agency (JST)
Apr. 2011
Associate Professor, Kyoto University, Institute for Integrated Cell-Material Sciences (iCeMS); Visiting Associate Professor, The National Centre for Biological Sciences (NCBS) / Institute for Stem Cell Biology and Regenerative Medicine (inStem)
Apr. 2017
Professor, Center for Highly Advanced Integration of Nano and Life Sciences (G-CHAIN), Gifu University
Jan. 2021
Professor, Cell Biophysics Laboratory of the Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System, Gifu University
Apr. 2023
Chief, Division of Advanced Bioimaging, National Cancer Center Research Institute (Concurrent)

Dr. Rinshi Kasai
Division of Advanced Bioimaging, National Cancer Center Research Institute: Laboratory Head.
Hoshi University: Visiting Senior Lecturer.
Mar. 2005
Department of Biological Science, School of Science, Nagoya University, Ph. D.
Apr. 2005
Researcher, International Cooperative Research Project (ICORP), Japan Science and Technology Agency
Apr. 2010
Researcher, Institute for Integrated Cell-Material Sciences (iCeMS), Kyoto University
Apr. 2011
Assistant Professor, Institute for Frontier Medical Sciences, Kyoto University
Oct. 2016
Assistant Professor, Institute for Frontier Life and Medical Sciences, Kyoto University
Apr. 2021
Program-Specific Associate Professor, Cell Biophysics Laboratory of the Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System, Gifu University
Jun. 2023
Laboratory Head, Division of Advanced Bioimaging, National Cancer Center Research Institute
Apr. 2024
Visiting Senior Lecturer, Hoshi University

Dr. Koichiro Hirosawa
Cell Biophysics Laboratory of the Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System, Gifu University: Researcher
Mar. 2010
Department of Micro Engineering, Kyoto University, Ph.D.
Apr. 2010
Researcher, Institute for Integrated Cell-Material Sciences (iCeMS), Kyoto University
Apr. 2017
Researcher, Center for Highly Advanced Integration of Nano and Life Sciences (G-CHAIN), Gifu University
Jan. 2021
Researcher, Cell Biophysics Laboratory of the Institute for Glyco-core Research (iGCORE), Tokai National Higher Education and Research System, Gifu University
Related product
The ORCA-Quest 2 is a new qCMOS® camera, the successor to the ORCA-Quest with further advances such as faster readout speeds in extremely low-noise scan mode and increased sensitivity in the ultraviolet region.
Other case studies
- Confirmation
-
It looks like you're in the . If this is not your location, please select the correct region or country below.
You're headed to Hamamatsu Photonics website for US (English). If you want to view an other country's site, the optimized information will be provided by selecting options below.
In order to use this website comfortably, we use cookies. For cookie details please see our cookie policy.
- Cookie Policy
-
This website or its third-party tools use cookies, which are necessary to its functioning and required to achieve the purposes illustrated in this cookie policy. By closing the cookie warning banner, scrolling the page, clicking a link or continuing to browse otherwise, you agree to the use of cookies.
Hamamatsu uses cookies in order to enhance your experience on our website and ensure that our website functions.
You can visit this page at any time to learn more about cookies, get the most up to date information on how we use cookies and manage your cookie settings. We will not use cookies for any purpose other than the ones stated, but please note that we reserve the right to update our cookies.
1. What are cookies?
For modern websites to work according to visitor’s expectations, they need to collect certain basic information about visitors. To do this, a site will create small text files which are placed on visitor’s devices (computer or mobile) - these files are known as cookies when you access a website. Cookies are used in order to make websites function and work efficiently. Cookies are uniquely assigned to each visitor and can only be read by a web server in the domain that issued the cookie to the visitor. Cookies cannot be used to run programs or deliver viruses to a visitor’s device.
Cookies do various jobs which make the visitor’s experience of the internet much smoother and more interactive. For instance, cookies are used to remember the visitor’s preferences on sites they visit often, to remember language preference and to help navigate between pages more efficiently. Much, though not all, of the data collected is anonymous, though some of it is designed to detect browsing patterns and approximate geographical location to improve the visitor experience.
Certain type of cookies may require the data subject’s consent before storing them on the computer.
2. What are the different types of cookies?
This website uses two types of cookies:
- First party cookies. For our website, the first party cookies are controlled and maintained by Hamamatsu. No other parties have access to these cookies.
- Third party cookies. These cookies are implemented by organizations outside Hamamatsu. We do not have access to the data in these cookies, but we use these cookies to improve the overall website experience.
3. How do we use cookies?
This website uses cookies for following purposes:
- Certain cookies are necessary for our website to function. These are strictly necessary cookies and are required to enable website access, support navigation or provide relevant content. These cookies direct you to the correct region or country, and support security and ecommerce. Strictly necessary cookies also enforce your privacy preferences. Without these strictly necessary cookies, much of our website will not function.
- Analytics cookies are used to track website usage. This data enables us to improve our website usability, performance and website administration. In our analytics cookies, we do not store any personal identifying information.
- Functionality cookies. These are used to recognize you when you return to our website. This enables us to personalize our content for you, greet you by name and remember your preferences (for example, your choice of language or region).
- These cookies record your visit to our website, the pages you have visited and the links you have followed. We will use this information to make our website and the advertising displayed on it more relevant to your interests. We may also share this information with third parties for this purpose.
Cookies help us help you. Through the use of cookies, we learn what is important to our visitors and we develop and enhance website content and functionality to support your experience. Much of our website can be accessed if cookies are disabled, however certain website functions may not work. And, we believe your current and future visits will be enhanced if cookies are enabled.
4. Which cookies do we use?
There are two ways to manage cookie preferences.
- You can set your cookie preferences on your device or in your browser.
- You can set your cookie preferences at the website level.
If you don’t want to receive cookies, you can modify your browser so that it notifies you when cookies are sent to it or you can refuse cookies altogether. You can also delete cookies that have already been set.
If you wish to restrict or block web browser cookies which are set on your device then you can do this through your browser settings; the Help function within your browser should tell you how. Alternatively, you may wish to visit www.aboutcookies.org, which contains comprehensive information on how to do this on a wide variety of desktop browsers.
5. What are Internet tags and how do we use them with cookies?
Occasionally, we may use internet tags (also known as action tags, single-pixel GIFs, clear GIFs, invisible GIFs and 1-by-1 GIFs) at this site and may deploy these tags/cookies through a third-party advertising partner or a web analytical service partner which may be located and store the respective information (including your IP-address) in a foreign country. These tags/cookies are placed on both online advertisements that bring users to this site and on different pages of this site. We use this technology to measure the visitors' responses to our sites and the effectiveness of our advertising campaigns (including how many times a page is opened and which information is consulted) as well as to evaluate your use of this website. The third-party partner or the web analytical service partner may be able to collect data about visitors to our and other sites because of these internet tags/cookies, may compose reports regarding the website’s activity for us and may provide further services which are related to the use of the website and the internet. They may provide such information to other parties if there is a legal requirement that they do so, or if they hire the other parties to process information on their behalf.
If you would like more information about web tags and cookies associated with on-line advertising or to opt-out of third-party collection of this information, please visit the Network Advertising Initiative website http://www.networkadvertising.org.
6. Analytics and Advertisement Cookies
We use third-party cookies (such as Google Analytics) to track visitors on our website, to get reports about how visitors use the website and to inform, optimize and serve ads based on someone's past visits to our website.
You may opt-out of Google Analytics cookies by the websites provided by Google:
https://tools.google.com/dlpage/gaoptout?hl=en
As provided in this Privacy Policy (Article 5), you can learn more about opt-out cookies by the website provided by Network Advertising Initiative:
http://www.networkadvertising.org
We inform you that in such case you will not be able to wholly use all functions of our website.
Close